Time: 2024-08-11
Non-small cell lung cancer Healthy Survival Prediction Tips
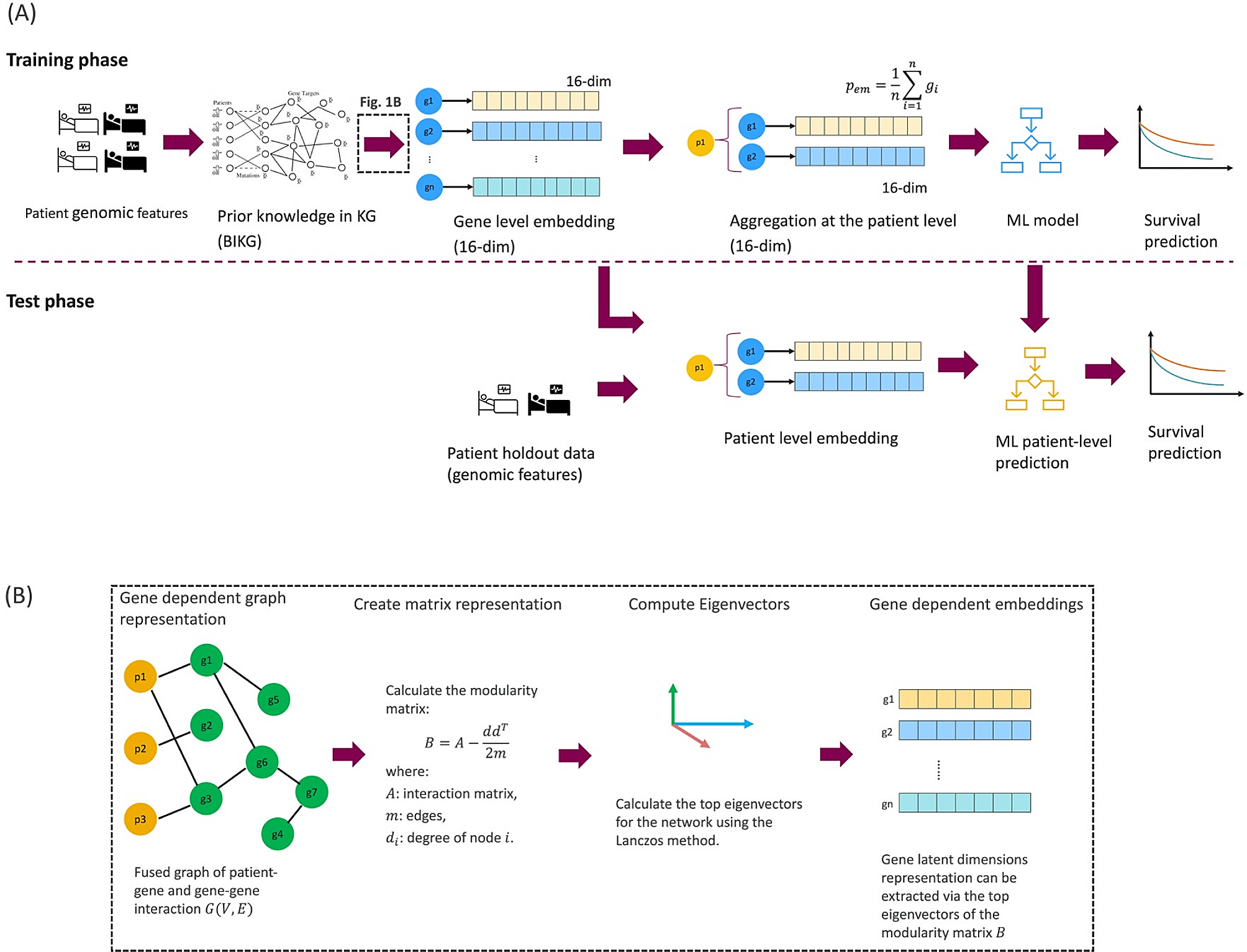
Prediction Framework for Patient Survival and Response
The framework for prediction of patient survival and response incorporates knowledge graphs ( KGs ) in patient survival prediction models . By using the patient 's genomic signature , gene information is projected to the KG , expanding knowledge to include gene - gene interactions . The identified connected graph is then projected to a lower - dimensional embedding using the SocioDim algorithm . Gene - specific embeddings are aggregated to a patient - level one for input into the machine learning ( ML ) patient survival prediction model . The use of various methods for generating low - dimensional embeddings from KGs was evaluated , with the SocioDim algorithm proving advantageous.
Integration of Patient Genomic Data with Knowledge Graphs
A stand - alone software application knowledge interface ( API ) was developed to incorporate patient genomic data with relevant prior knowledge from KGs as input to the ML model for predicting patient survival . The study compared the performance of internally developed knowledge graph BIKG with a well - established biomedical KG Hetionet , demonstrating the adaptability of the framework to various KG platforms.
Improvements in Prediction of Survival in NSCLC Patients
The proposed framework was applied to analyze improvements in using KGs to predict survival in non - small cell lung cancer ( NSCLC ) patients from different studies and clinical trials . The study evaluated multiple ML - based predictive algorithms for patient survival , with random survival forest proving to be the fastest , most stable , and most generalizable method.
Enhancing OS Prediction in NSCLC Patients
The accuracy of OS prediction was enhanced through the incorporation of prior knowledge from KGs in the analysis of NSCLC patients treated with immune checkpoint inhibitors ( IO ) in clinical trials . Models trained with the BIKG in combination with gene panel data outperformed models based solely on gene panel data in predicting OS in both OAK and POPLAR clinical trials.
Identification of Biomarker - Based Signature
The study identified a biomarker - based signature for differentiating OS in NSCLC patients using a combination of key genes . The gene mutation signature as an OS differentiator was found to significantly differentiate OS in patient cohorts from the OAK and MSK studies . The prevalence of mutations in predicted high- and low - risk groups was compared , with models using the BIKG identifying more high - risk patients associated with specific gene mutations.
Overall , the incorporation of prior knowledge from KGs in predictive models shows promise in improving the accuracy of OS prediction in NSCLC patients , particularly when combined with gene panel data . The study highlights the potential of using biomarker - based signatures to differentiate OS outcomes in NSCLC patients across various datasets and clinical trials.
-
HealthTime: 2024-08-11Discover the role of dopamine in social drinking and its impact on alcohol consumption . Learn how social settings amplify euphoric feelings and the implications for Alcohol Use Disorder . Join thousands in understanding the neurobiological processes behind social drinking .
-
HealthTime: 2024-08-10Discover the preventive measures taken to combat the Chandipura virus outbreak in Gujarat , including surveillance activities and spraying of pesticide . Learn how medical experts are addressing the symptoms of the virus and the efforts to control its spread . Join thousands of health officials in raising awareness and preventing further infections in affected areas .
-
HealthTime: 2024-08-10Discover the risk of variant H3N2 flu cases in Ingham County and learn how to prevent and respond to the virus . Get instant tips for healthy wellness and stay informed to mitigate the impact of H3N2 flu infections .
-
HealthTime: 2024-08-10Discover the guaranteed wellness tips to protect yourself from the West Nile virus in Centre County . Learn how to stay healthy and safe with these essential mosquito protection tips .
-
HealthTime: 2024-08-10Discover the potential of classical music as a complementary tool in depression treatment . Learn how personalized music therapy plans can enhance treatment outcomes and improve mental health . Join thousands of individuals in leveraging music for managing emotions and improving overall well - being .
-
HealthTime: 2024-08-10Discover the mental health benefits of a Mediterranean diet in reducing stress levels . Learn how incorporating whole grains , seafood , fruits , and vegetables can positively impact your psychological well - being . Start improving your mental wellness today with these diet tips .


 Business
Business